Peptide Identification for MHC-binding Peptidome
Introduction:
Peptides bound to cell surface MHC molecules allow the immune system to recognize intracellular pathogens and tumor-derived peptides. Each MHC allele has a distinct peptide binding motif which favours certain amino acids at particular points in a sequence.
Omics Technologies scientists developed a way that could capture essentially all MHC molecules and could harvest the peptides presented by those MHC molecules. Through in-depth proteome analysis and large scale database search, including numerous cancer mutation databases, we could give you a complete profile of your MHC-binding peptidome.
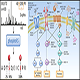
There are typically 9 steps in a MHC-binding Peptidomics Analysis project:
1) MHC Capturing – Numerous methods are applied to enrich MHC molecules in your sample and MHC complexes are harvested through elution using My-IPElu™ buffers.
2) Sample preparation – Cell or tissue lysis, biofluid (plasma or serum) depletion; protein quantification on all samples.
3) Sample digestion – Reduction and alkylation followed by in-solution trypsin digestion and C18 cleaning before labeling.
4) SCX cleaning – Ion-exchange to remove excessive chemicals and fractionation of pooled sample through off-line SCX HPLC.
5) 2D fractionation – bRPLC separation of the SCX cleaned samples into 96 fractions, and further pooled into 24 fractions based on the rule of avoiding co-elution.
6) Nano LC/MS/MS – 50 nL/min liquid chromatography to enable super high ionization efficiency for downstream Mass Spectrometry Analysis using the state-of-the-art Orbitrap Mass Spectrometer. 24 fractions will be analyzed in 24 Mass Spectrometry runs.
7) Protein profiling – Protein database searching with Mascot.
8) Data analysis – Data validation, visualization and quantification using commercial softwares and Omics Technologies's unique R packages and scripts.
9) Report generation – Report will be sent to you in Excel format as well as a summary in PDF format. We will also provide you any details you need for your papers' MATERIALS AND METHODS section. We will make sure you understand your result and help you with your paper writing with free follow-up services.
Sample types we accept:
1, 2D gel spots, SDS-PAGE bands
2, Cell Lysates and Tissue Lysates
3, Biofluids, such as plasma*, serum*, saliva, tear, etc.
*We provide High Abundance Protein Depletion Service to significantly (100-500 folds) increase the depth of your biofluid proteomics analysis by removing top abundant proteins from your samples. Read more for details.
4, FFPE slide/ FFPE extract
5, Customized sample types (please contact us to discuss)
Please refer to our Omics Technologies Proteomics Sample Preparation and Shipping Guidelines for instructions on how to prepare your samples with less contamination for Mass Spectrometry Analysis.
-
Phone:
+1 888-206-0066 (TOLL FREE) -
Email:
info@omicstech.com